Maria Cristina Gambetta received a joint PhD in 2010 from the EMBL in Heidelberg (Germany) and the University of Geneva (Switzerland), and performed post-doctoral research at the Max-Planck Institute of Biochemistry in Martinsried (Germany), studying molecular mechanisms of epigenetic gene silencing by Polycomb transcriptional repressors under the supervision of Jürg Müller. During her post-doctoral training at the EMBL in the laboratory of Eileen Furlong, she studied the role of chromatin architecture in gene activation. She joined the Center for Integrative Genomics in January 2018 as Assistant Professor to study how cells organize the activating and repressive regulatory inputs that are necessary to achieve appropriate gene expression patterns. She was promoted to Associate Professor in August 2023.
For more information please visit: https://gambettalab.org
Research summary
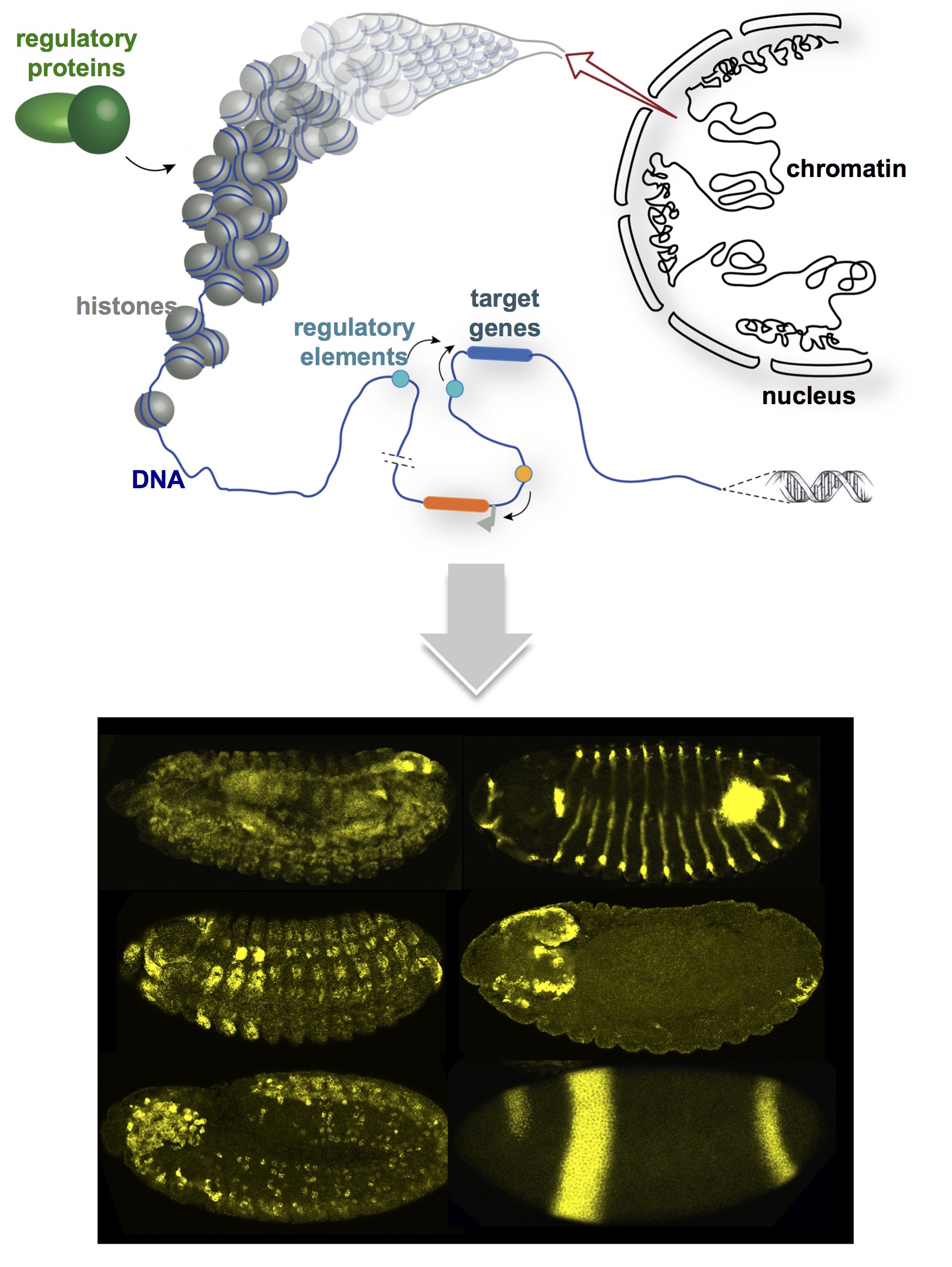
In multicellular organisms, genes are turned ON and OFF in robust spatial and temporal patterns. Regulatory elements that activate or silence gene transcription are inherently promiscuous and can act over long genomic distances. A basic question is how regulatory elements find their target promoters and restrict their activity to their targets.
On the one hand, we study how regulatory crosstalk is prevented. Insulators were discovered in Drosophila and subsequently in various organisms from yeast to humans, as DNA elements that block communication between a regulatory element and a gene promoter when placed in between. In flies and humans, insulators exert their activity by recruiting insulator proteins. We study the biological relevance and molecular mechanisms of insulator proteins.
On the other hand, we study how genes find their appropriate regulatory elements across surprisingly long genomic distances and skip over non-target genes. We address the biological relevance of these interactions by re-wiring them, and we study how they form.
Our studies in flies reveal new evolutionary perspectives into the relevance of 3D genome folding for correctly wiring genes to their regulatory elements.
Masters, PhD and Postdoc positions available; people interested in joining the team are encouraged to write directly to mariacristina.gambetta@unil.ch.
Publications
 |
Pascal Cousin 2005 to 2019 - Technician, Center for Integrative Genomics, University of Lausanne, Switzerland |
 |
Anastasiia Semenova 2017 to 2019 - M.Sc. Biology, Taras Shevchenko National University of Kyiv, ESC Institute of Biology and Medicine, Kyiv, Ukraine 2013 to 2017 - B.Sc. Biology, Taras Shevchenko National University of Kyiv, ESC Institute of Biology and Medicine, Kyiv, Ukraine |
 |
Bihan Wang 2017 to 2018 - M.Sc. Biochemistry, University of Florida, USA 2010 to 2014 - B.Sc. Wuhan University, School of Pharmaceutical Science, China |
 |
Marion Mouginot 2018 to 2020 - M.Sc. Cellular and Molecular Biology, Lyon 1 University, France 2016 to 2018 - Assistant engineer, École Normale Supérieure of Lyon (ENSL), France 2015 to 2016 - B.Sc. Biotechnology speciality Genomics, Lyon 1 University, France 2013 to 2015 - Technical degree in Biotechnology, Martinière Duchère high school, France |
 |
Jenisha Khadka 2020 to 2022 - Research Intern, Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany 2018 to 2021 - M.Sc. in Molecular Life Sciences, Friedrich-Schiller-Universität Jena, Jena, Germany 2013 to 2017 - B.Sc. in Biotechnology, Purbanchal University, Kathmandu, Nepal |
 |
Sanyami Sunil Zunjarrao 2019 to 2021 - Project Assistant, National Chemical Laboratory, India 2014 to 2019 - M.Sc., Institute of Bioinformatics and Biotechnology, Savitribai Phule Pune University, India |
 |
Sahar Hani 2022 to 2023 - Postdoctoral fellow, Leibniz Institute für Gemüse- und Zierpflanzenbau (IGZ), Grossbeeren, Germany 2018 to 2022 - PhD Plant Molecular Biology, Aix-Marseille Institute of Biosciences and Biotechnologies, CEA Cadarache, France 2016 to 2018 - M.Sc. Applied Plant Biotechnology, Faculty of Sciences, Lebanese University, Lebanon 2013 to 2016 - B.Sc. Biology, Faculty of Sciences, Lebanese University, Lebanon |

