Biography
Andreas Mayer studied biology and chemistry at the University of Munich. In 1995 he obtained a PhD from the same university for studies on protein translocation into mitochondria in the laboratory of Walter Neupert. After postdoctoral studies on organelle inheritance and fusion with William Wickner at Dartmouth Medical School, Andreas Mayer joined the Friedrich-Miescher-Laboratorium of the Max-Planck-Society as a group leader in 1997. In 2003, he joined the University of Lausanne as full professor of biochemistry.
Research Interests
1. Membrane fission on endo-lysosomal compartments
Endo-lysosomal compartments control the abundance of most plasma membrane transporters and receptors, and thereby determine the communication, interaction and transport capacities of cells. Endosomes receive proteins from the cell surface or from the Golgi and recycle them either back to these compartments, or transfer them to lysosomes, where many of them become degraded. Endo-lysosomal compartments vigorously exchange material through homo- and heterotypic fusion and fission, and through tubulo-vesicular carriers. Carriers grow by polymerisation of proteinaceous coats, such as retromer, which collect cargo proteins and then detach from the organelle by membrane fission.
Persistence of the endo-lysosomal network depends on the control of the formation and fission of such carriers and on their coordination with the availability of membrane and/or cargo. It has remained enigmatic how it this coordination is achieved.
We address this question using lysosome-like yeast vacuoles as a highly versatile model for discovery and utilize the better developed endo-lysosomal system of mammalian cells for further in-vivo analyses and for testing evolutionary conservation. The mechanisms of retromer-dependent formation and fission of membrane carriers are explored by in-vitro systems, such as synthetic lipid tubules and giant unilamellar vesicles, which are combined with in vivo analyses to test the relevance of the results for the endo-lysosomal system of eukaryotic cells.
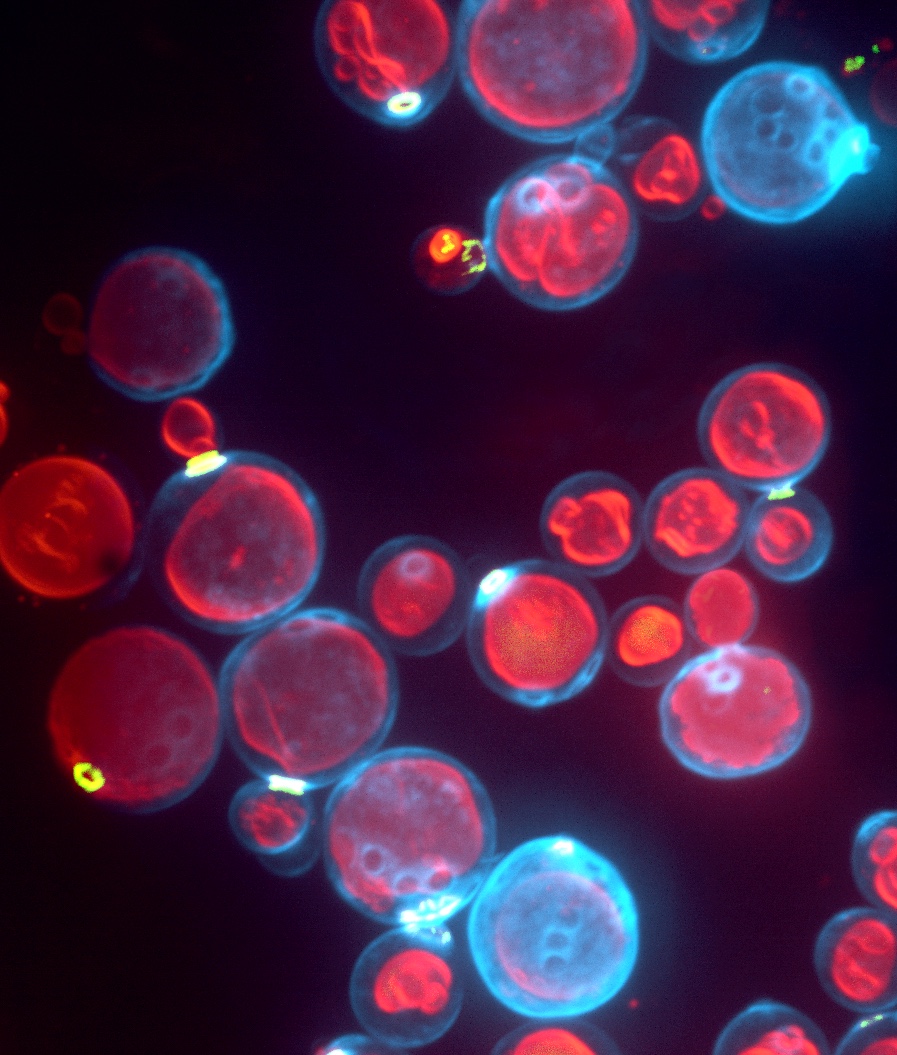 |
Fig. 1: Systems to study coat formation and membrane fission.
Left: Retromer coats growing on oriented lipid tubules in a microfluidic chamber. Retromer subunits colored in red and green, lipid in blue. Right: Lysosome-like vacuoles in living yeast cells. Cell walls are stained in blue, vacuoles in red.
2. The INPHORS pathway: A novel signaling pathway regulating phosphate homeostasis in the cytosol
Cells actively manage the concentration of free, inorganic phosphate (Pi) in their cytosol. Pi homeostasis is crucial because cells must strike a balance between conflicting goals. On the one hand, Pi is a product of nucleotide-hydrolyzing reactions. Its concentration has a significant impact on the free energy that these reactions can provide to drive metabolism. Since, for his reason, excessive Pi concentrations might stall metabolism, cytosolic Pi must remain limited. On the other hand, Pi is an essential macronutrient. As a major constituent of nucleic acids, phospholipids and carbohydrates, it is consumed in large quantities for anabolic reactions. Aside from this vital importance for metabolism, Pi homeostasis is of interest for agriculture, because Pi supply is limiting for crop yields. Its medical relevance is underlined by mutations perturbing intracellular phosphate homeostasis, which lead to neurodegenerative diseases or may perturb Pi resorption in the kidney.
To maintain the critical intracellular balance of Pi, cells dispose of a dedicated signaling pathway for intracellular phosphate reception and signaling (INPHORS) and of a buffering system in the form of acidocalcisomes. We discovered that inositol pyrophosphates, compounds of so far poorly understood functions, communicate cytosolic Pi levels to SPX domains, a family of conserved protein modules of hitherto unknown function (Wild et al., Science 352, 986; Gerasimaite et al., ACS Chem. Bio. 12, 648). SPX domains bind or are linked to many proteins for uptake, excretion or storage of Pi, or for transcriptional phosphate starvation responses. Our results thus provided first evidence for a novel pathway dedicated to Pi homeostasis, and they identified SPX domains and inositol pyrophosphates as its first two elements. We now seek to elucidate this pathway entirely.
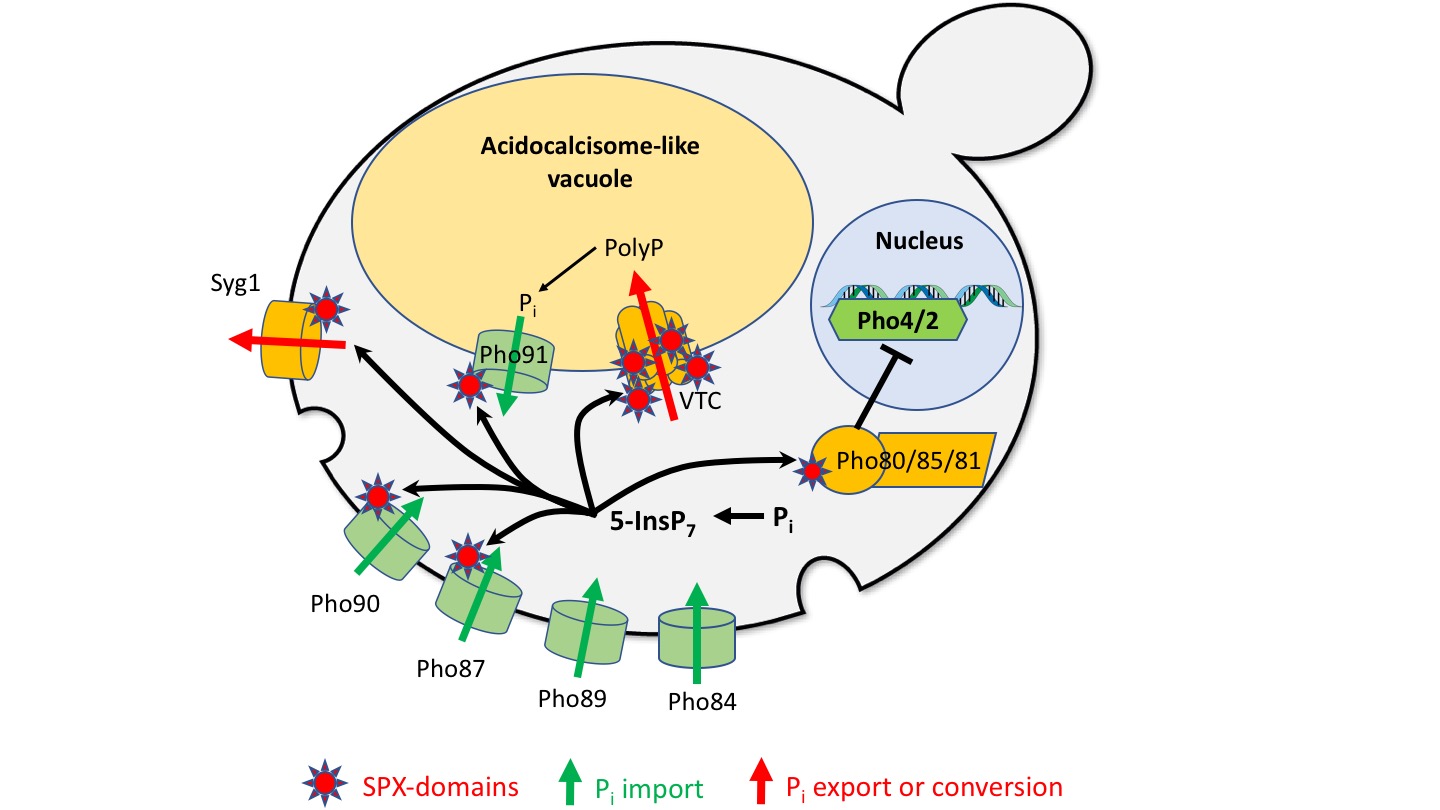
Fig. 2: System for Pi homeostasis in the cytosol of yeast cells.
Pi can be imported into the cytosol by high-affinity (Pho89/84) and low affinity importers (Pho87/90) and by a Pi-permeable vacuolar channel (Pho91). It can be exported from the cytosol by secretion into the extracellular space (Syg1) or by conversion into polyP and transfer into the acidocalcisome-like vacuole (VTC). Inside this organelle, polyphosphatases can re-convert polyP into Pi to make it available for export into the cytosol. A Pi-regulated kinase (Pho80/85/81) phosphorylates transcription factors (Pho4/2) for the transcriptional phosphate starvation response. A sensor for cytosolic Pi might regulate the levels of inositol pyrophosphates (e.g. 5-InsP7), which then act on SPX domains. These should, when bound to inositol pyrophosphates, inactivate proteins importing Pi and activate proteins exporting Pi from the cytosol.
Cells dispose of a buffering system for intracellular Pi, constituted by acidocalcisomes or acidocalcisome-like vacuoles, highly conserved lysosome-related organelles that are present from bacteria to men, but the mechanisms by which they exert these functions are poorly understood. These organelles convert cytosolic Pi into inorganic polyphosphate, chains of dozens or hundreds of Pi residues linked by phosphoric anhydride bonds. Acidocalcisomes store this compound to high concentrations in their lumen and can reconvert it into Pi upon demand. We explore how the conversion and release of Pi by acidocalcisomes is controlled by cytosolic Pi and inositol pyrophosphates, such that these organelles guarantee cytosolic Pi homeostasis.
| Person | Position | Contact |
|---|---|---|
| Véronique Comte-Miserez | Technician | Unisciences |
| Maria Giovanna De Leo | Postdoctoral Fellow | Unisciences |
| Navin Gopaldass | Research Associate | Unisciences |
| M. Jamsheer Kaithakkal | Postdoctoral Fellow | Unisciences |
| Geundon Kim | Postdoctoral Fellow | Unisciences |
| Matthias Künzle | Postdoctoral Fellow | Unisciences |
| Lydie Michaillat Mayer | Research Associate | Unisciences |
| Ana Catarina (Rodrigues) Alves | Postdoctoral Fellow | Unisciences |
| Sudeshna Roy Chowdhury | Postdoctoral Fellow | Unisciences |
| Andrea Schmidt Luther | Technician | Unisciences |

