Single pathway targeted analyses (absolute quantification)
| alanine | glutamate | ornithine |
| alpha-aminoadipate | glutamine | phenylalanine |
| alpha-aminobutyrate | glycine | pipecolate |
| anserine | guanidinoacetate | proline |
| arginine | histidine | serine |
| asparagine | homocitrulline | taurine |
| beta-alanine/sarcosine | isoleucine | threonine |
| carnosine | kynurenine | trans-4-hydroxy-proline |
| citrulline | leucine | tryptophan |
| creatine | lysine | tyrosine |
| gamma-aminobutyrate | methionine | valine |
| Carnitine |
| Acetylcarnitine |
| Propionylcarnitine |
| Iso-/Butyrylcarnitine |
| Isovaleryl-/2-Methylbutyrylcarnitine |
| Hexanoylcarnitine |
| Octanoylcarnitine |
| Decanoylcarnitine |
| Dodecanoylcarnitine |
| Tetradecanoylcarnitine |
| Palmitoylcarnitine |
| Stearoylcarnitine |
Other hydroxy- and (un)saturated acylcarnitinespecies can also be measured on demand:
| 3-Hydroxy-butyrylcarnitine | Heptanoylcarnitine |
| 3-OH-decanoylcarnitine | Hexadecanoylcarnitine |
| 3-OH-dodecanoylcarnitine | Hexadecenoylcarnitine |
| 3-OH-hexadecanoylcarnitine | Hydroxyisovalerylcarnitine |
| 3-OH-hexadecenoylcarnitine | Linoleylcarnitine |
| 3-OH-hexanoylcarnitine | Malonylcarnitine |
| 3-OH-linoleylcarnitine | Methylmalonylcarnitine |
| 3-OH-oleylcarnitine | Octenoylcarnitine |
| 3-OH-stearoylcarnitine | Oleylcarnitine |
| 3-OH-tetradecanolycarnitine | Suberylcarnitine, propylglutarylcarnitine |
| 3-OH-tetradecenoylcarnitine | Succinylcarnitine |
| Adipylcarnitine, 3-methylglutarylcarnitine | Tetradecadienoylcarnitine |
| Decenoylcarnitine | Tetradecenoylcarnitine |
| Dodecenoylcarnitine | Tiglylcarnitine |
| Glutarylcarnitine |
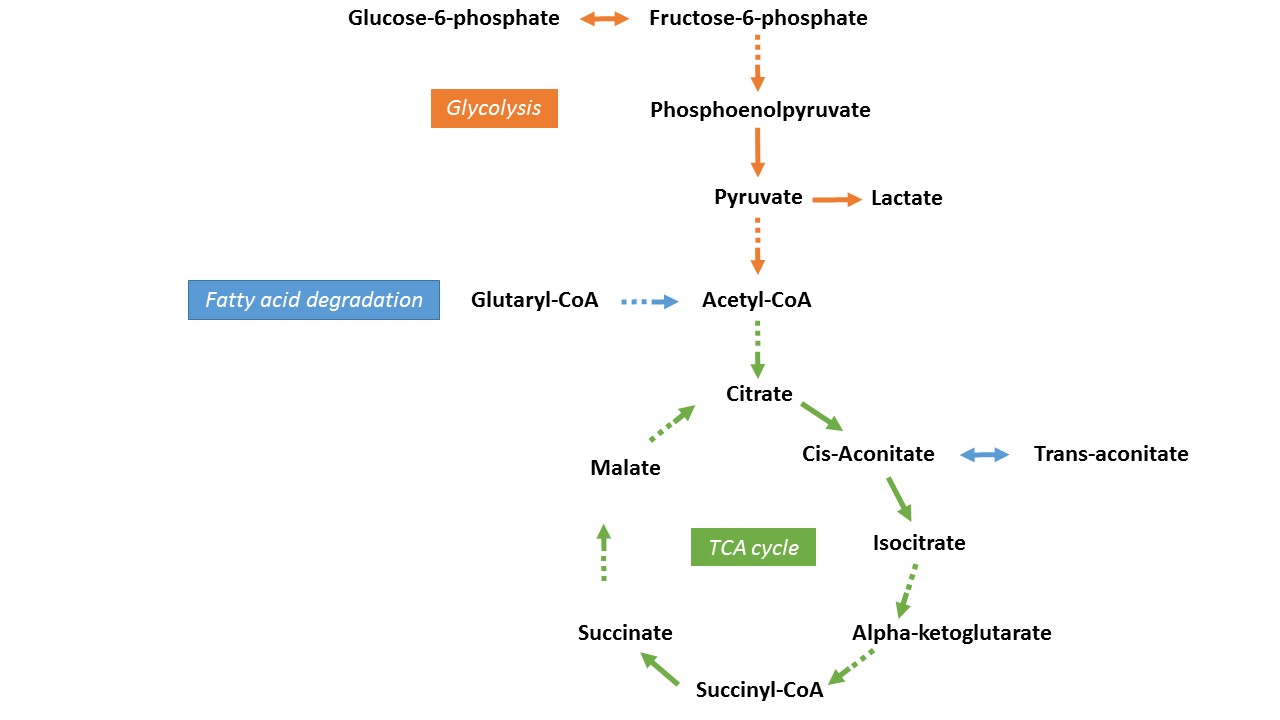
| Acetyl-CoA | Glutaryl-CoA |
| Citrate | Trans-aconitate |
| Isocitrate | Glucose-6-phosphate |
| Alpha-ketoglutarate | Fructose-6-phosphate |
| Succinyl-CoA | Phosphoenolpyruvate |
| Succinate | Pyruvate |
| Malate | Lactate |
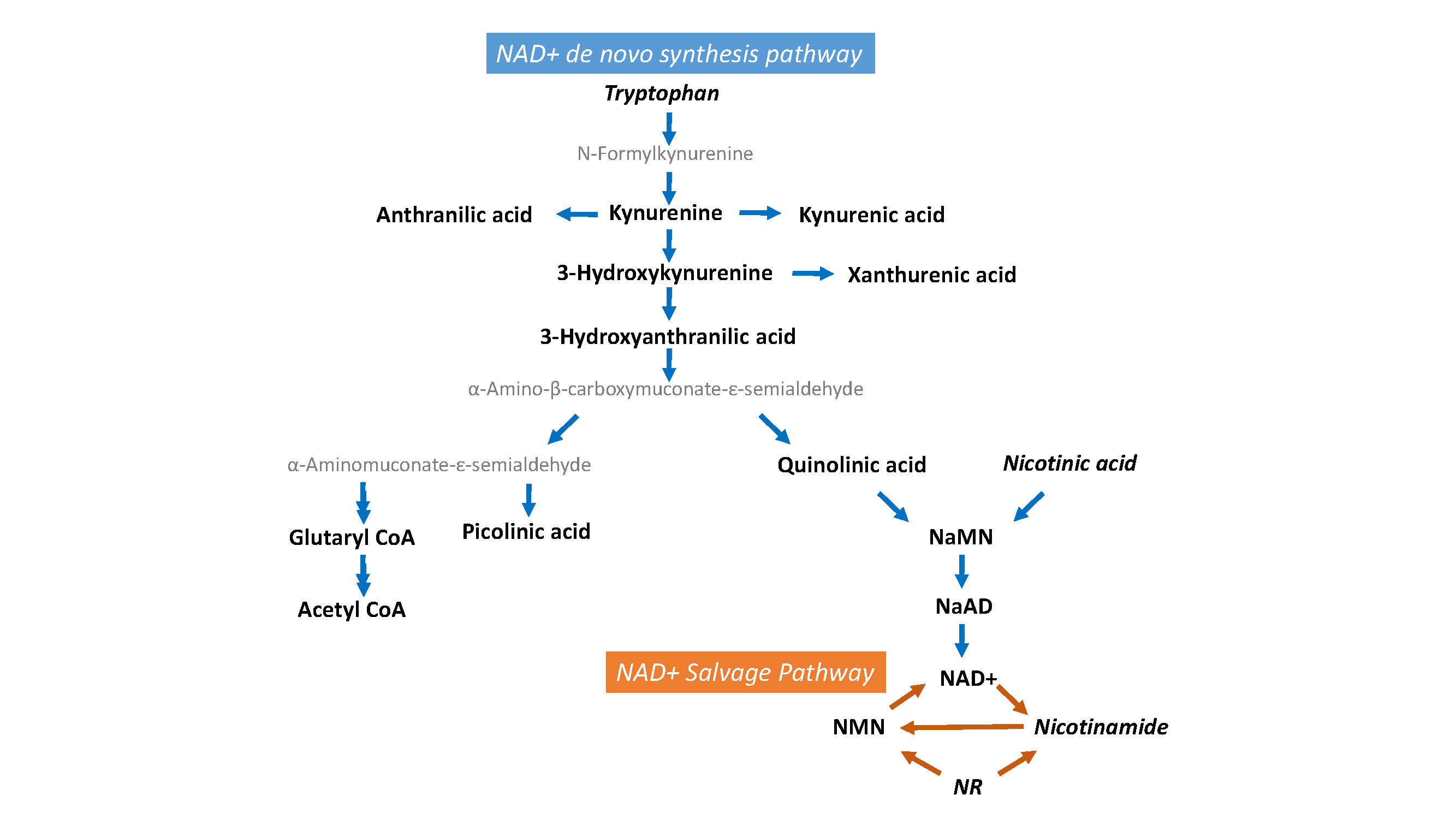
| tryptophan | nicotinic acid mononucleotide (NaMN, nicotinate ribonucleotide) |
| kynurenine | nicotinic acid adenine Dinucleotide (NaAD, deamino-NAD+) |
| kynurenic acid | NAD+ |
| anthranilic acid | nicotinamide mononucleotide (NMN, nicotinamide ribotide) |
| 3-hydroxy-kynurenine | nicotinamide riboside (NR, ribosyl-nicotinamide) |
| xanthurenic acid | nicotinamide (NAM, niacin, Vitamin B3) |
| 3-hydroxyanthranilic acid | picolinic acid |
| quinolinic acid | |
| nicotinic acid |
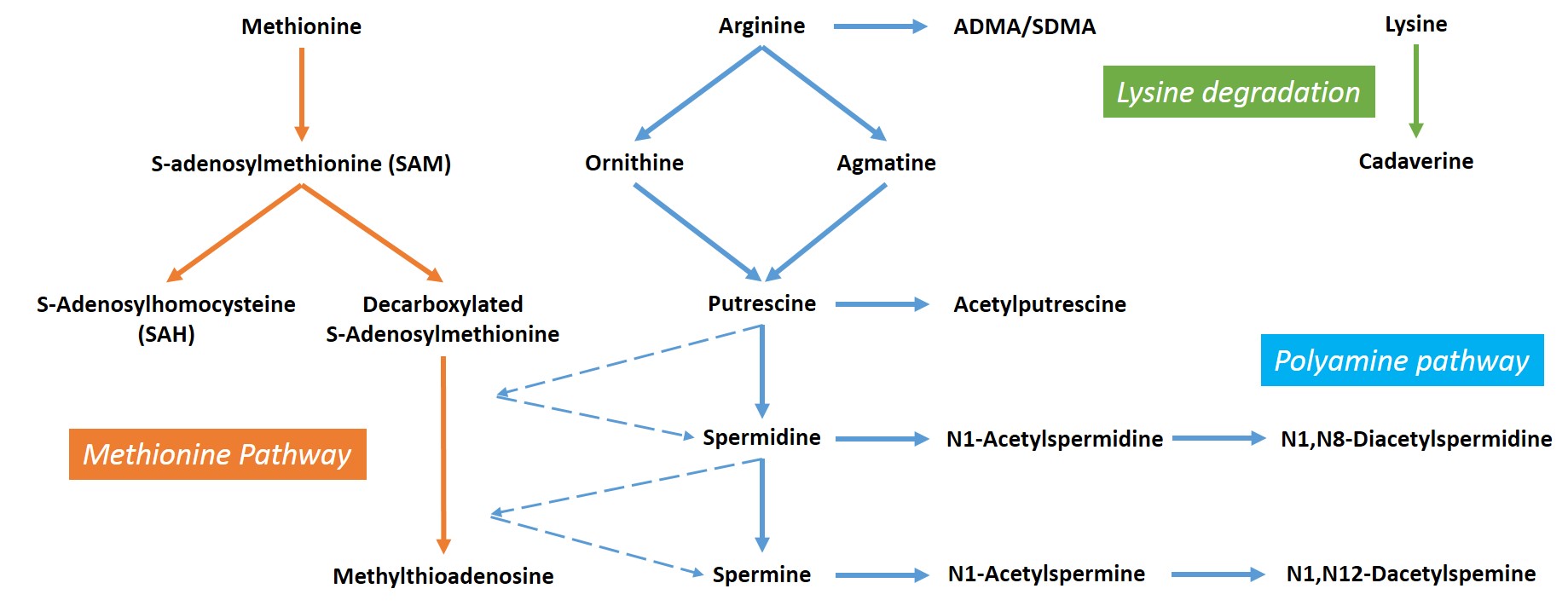
Methionine
S-adenosylmethionine (SAM)
S-adenosylhomocysteine (SAH)
Decarboxylated S-adenosylmethionine
Methylthioadenosine
Arginine
Asymmetric dimethylarginine (ADMA)
Symmetric dimethylarginine (SDMA)
Agmatine
Ornithine
Putrescine
Spermidine
Spermine
Acetylputrescine
N1-Acetylspermine
N1-Acetylspermidine
N1, N8-Diacetylspermidine
N1, N12-Diacetylspermine
Lysine
Cadaverine
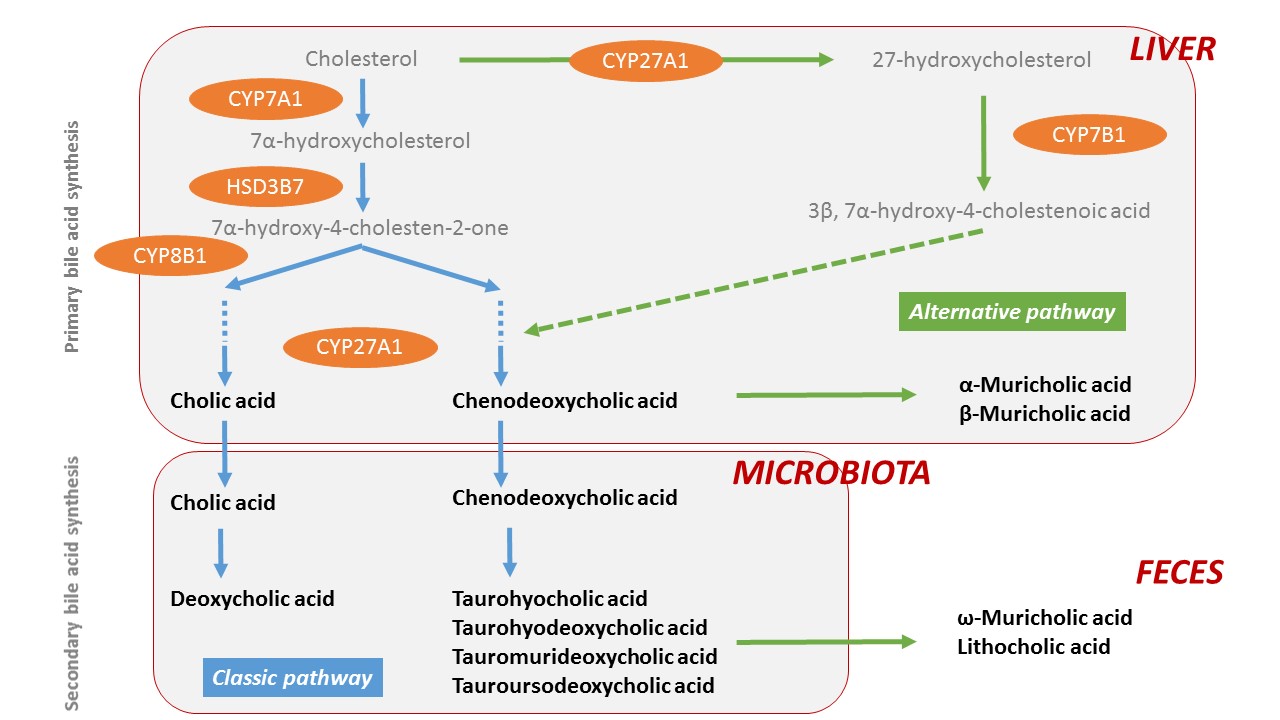
|
Lithocholic acid |
Cholic acid |
|
7-Ketolithocholic acid |
Glycolithocholic acid |
|
Murocholic acid |
Glycoursodeoxycholic acid |
|
Ursodeoxycholic acid |
Glycohyodeoxycholic acid |
|
Hyodeoxycholic acid |
Glycochenodeoxycholic acid |
|
Chenodeoxycholic acid |
Glycodeoxycholic acid * |
|
Deoxycholic acid |
Glycocholic acid |
|
Isodeoxycholic acid |
Taurolithocholic acid |
|
3-Oxocholic acid |
Tauroursodeoxycholic acid |
|
7-Ketodeoxycholic acid |
Taurohyodeoxycholic acid |
|
α-Muricholic acid |
Taurochenodeoxycholic acid |
|
β-Muricholic acid |
Taurodeoxycholic acid |
|
ω-Muricholic acid |
Tauro-β-muricholic acid |
|
γ-Muricholic acid |
Taurocholic acid |
| Progestagens | Androgens |
| Pregnenolone | Dehydroepiandrosterone |
| 17α-hydroxypregnenolone | Androstenedione |
| Progesterone | Testosterone |
| 17α-hydroxyprogesterone | Dihydrotestosterone |
| Glucocorticoids and mineralocorticoids | Androsterone |
| 11-deoxycorticosterone | EpiTestosterone |
| Corticosterone | Estrogens |
| Aldosterone | Estrone |
| 11-deoxycortisol | Estradiol |
| Cortisol | Estriol |
| 21-Deoxycortisol | EthynylEstradiol |
| Cortisone | Sulfated forms |
| Estrone sulfate | |
| DHEAS | |
| Pregnenolone sulfate |
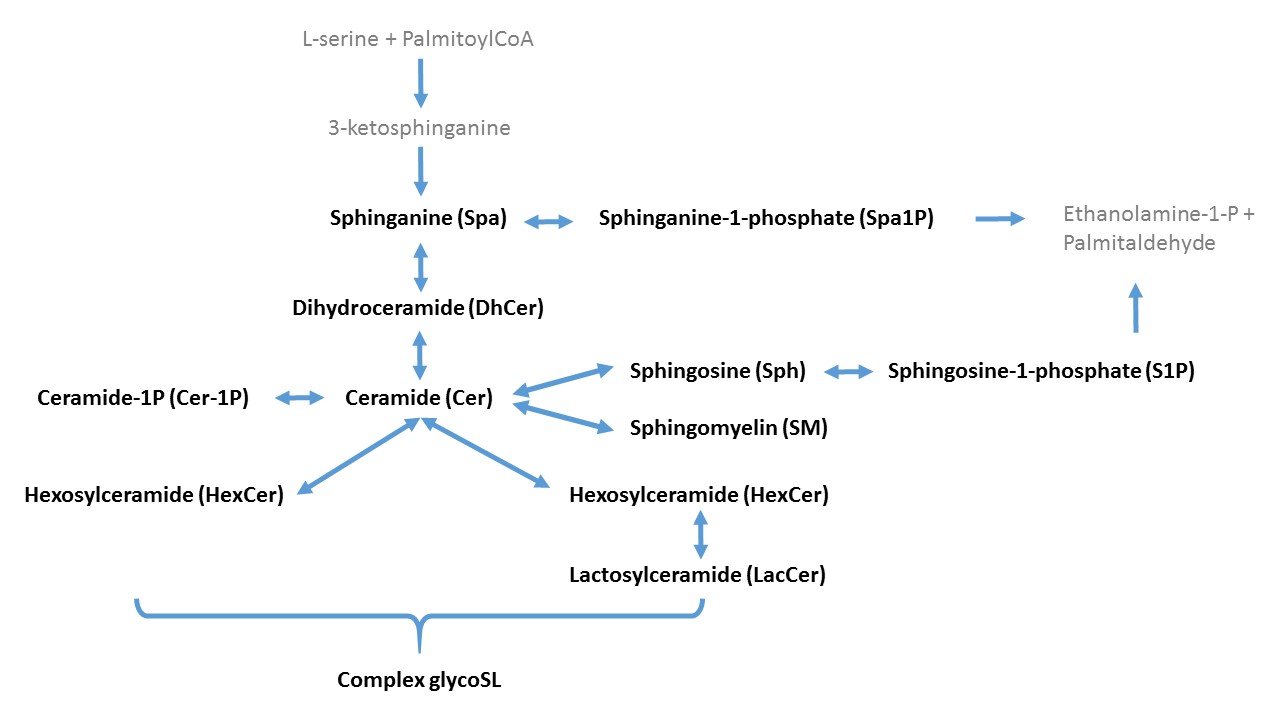
Method was recently updated to include deoxysphingolipids!
|
SM C6:0 (d18:1/6:0) |
DhCer C16:0 (d18:0/16:0) |
|
SM C12:0 (d18:1/12:0) |
DhCer C24:1 (d18:0/24:1(15Z)) |
|
SM C16:0 (d18:1/16:0) |
DhCer C24:0 (d18:0/24:0) |
|
SM C18:1 (d18:1/18:1(9Z)) |
GlcCer C12:0 (d18:1/12:0) |
|
SM C18:0 (d18:1/18:0) |
GlcCer C16:0 (d18:1/16:0) |
|
SM C24:1 (d18:1/24:1(15Z)) |
GlcCer C18:1 (d18:1/18:1(9Z)) |
|
SM C24:0 (d18:1/24:0) |
GlcCer C18:0 (d18:1/18:0) |
|
Cer C2:0 (d18:1/2:0) |
GlcCer C24:1 (d18:1/24:1(15Z)) |
|
Cer C4:0 (d18:1/4:0) |
LacCer C12:0 (d18:1/12:0) |
|
Cer C6:0 (d18:1/6:0) |
LacCer C16:0 (d18:1/16:0) |
|
Cer C8:0 (d18:1/8:0) |
LacCer C24:1 (d18:1/24:1) |
|
Cer C10:0 (d18:1/10:0) |
LacCer C24:0 (d18:1/24:0) |
|
Cer C12:0 (d18:1/12:0) |
GlcSph C18:1 |
|
Cer C14:0 (d18:1/14:0) |
Sph (d18:1) |
|
Cer C16:0 (d18:1/16:0) |
Sph (d20:1) |
|
Cer C18:1 (d18:1/18:1(9Z)) |
Spa (d18:0) |
|
Cer C18:0 (d18:1/18:0) |
Sph 1P (d18:1) |
|
Cer C20:0 (d18:1/20:0) |
Spa 1P (d18:0) |
|
Cer C22:0 (d18:1/22:0) |
Cer1P C16:0 (d18:1/16:0) |
|
Cer C24:1 (d18:1/24:1(15Z)) |
Cer1P C18:1 (d18:1/18:1(9Z)) |
|
Cer C24:0 (d18:1/24:0) |
Cer1P C24:0 (d18:1/24:0) |
|
13-HODE |
20-COOH-LTB4 |
5,6-DiHETE |
|
9-HODE |
5,6-DiHETrE |
5-KETE |
|
12(13)-EpOME |
15-KETE |
8(9)-EpETrE |
|
12,13-DiHOME |
5-HEPE |
9-HOTrE |
|
12-HETE |
8-iso-PGE2 |
8-HDoHE |
|
15-HETE |
LTB3 |
9-KOTrE |
|
5-HETE |
LTB4 |
20-OH-LTB4 |
|
9(10)-EpOME |
PGE1 |
13-HOTrE |
|
9,10,13-TriHOME |
6-keto-PGF1a |
8,15-DiHETE |
|
9,10-DiHOME |
PGD2 |
LTC4 |
|
9,12,13-TriHOME |
PGD3 |
14,15-LTC4 |
|
9-HETE |
PGE2 |
LTD4 |
|
13-KODE |
10,17-DiHDoHE |
LXA4 |
|
9-KODE |
11-HDoHE |
LXA5 |
|
12-HEPE |
δ 12-PGJ2 |
LXB4 |
|
15-HEPE |
17(R)-Resolvin-D1 |
18-HEPE |
|
14-HDoHE |
11(12)-EpETrE |
7,17-hydroxy DPA |
|
17-HDoHE |
PGF2a |
7-Maresin-1 |
|
14,15-DiHETE |
11,12-DiHETrE |
tetranor-PGDM |
|
EKODE |
14,15-DiHETrE |
13,14-dihydro-15-keto PGE2 |
|
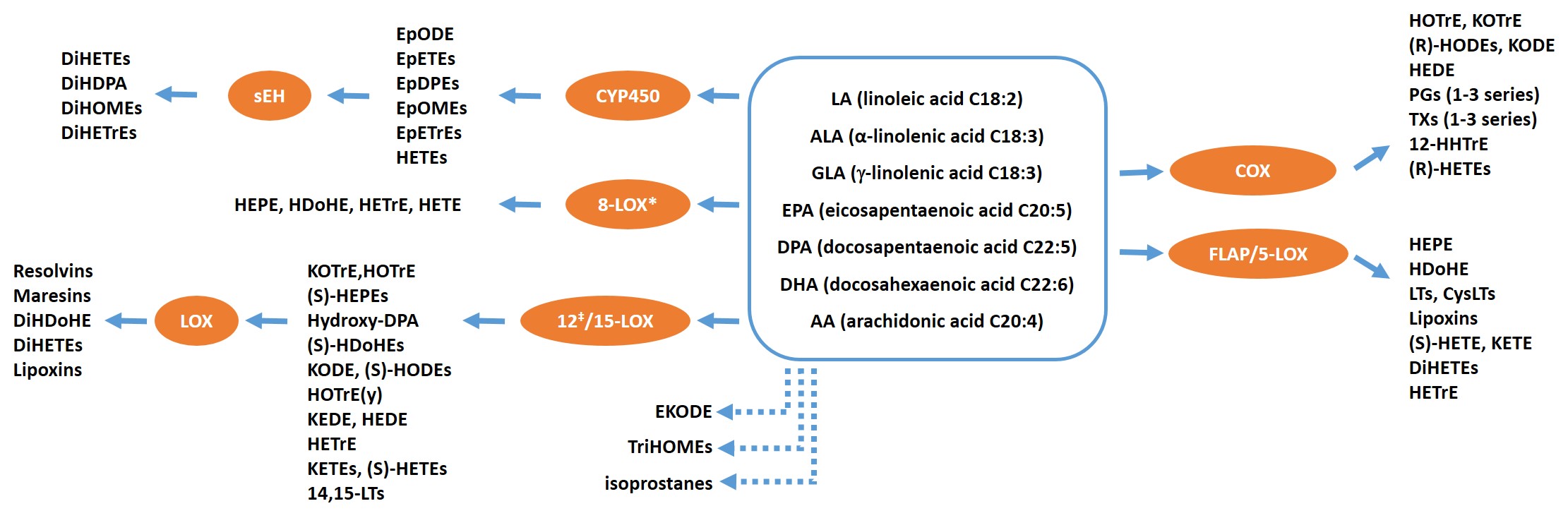
Arachidonic acid |
PGE3 |
tetranor-PGEM |
|
Linoleic Acid |
PGJ2 |
iPF2α-IV |
|
Docosahexaenoic Acid |
Resolvin D1 |
5-iPF2α-VI |
|
Eicosapentaenoic Acid |
Resolvin D2 |
8-iso-PGF2α |
|
TXB2 |
12(13)-EpODE |
8-iso-PGF3α |
|
11B-PGF2a |
12(S)-HHTrE |
11-dehydro TXB2 |
|
15-deoxy-PGJ2 |
LTE4 |
TXB3 |
|
19,20-DiHDPA |
5-HETrE |
11-dehydro TXB3 |
|
8-HETE |
14(15)-EpETE |
4-HDoHE |
|
11-HEPE |
14(15)-EpETrE |
11-HEDE |
|
PGB2 |
6-trans-LTB4 |
15-HEDE |
|
PGD1 |
8,9-DiHETrE |
13-HOTrE(γ) |
|
12-KETE |
16(17)-EpDPE |
15-epi Lipoxin A4 |
|
9-HEPE |
17(18)-EpETE |
TXB1 |
|
20-HETE |
17,18-DiHETE |
FOG9 |
|
8-HEPE |
19(20)-EpDPE |
19-HETE |
|
11-HETE |
5(6)-EpETrE |
15-OxoEDE |
|
8-HETrE |
5,15-DiHETE |
14,15-LTE4 |
|
15(S)-HETrE |
|
|
Method under development
Cholesterol
7a, 25-dihydroxycholesterol (7a25-OHC)
7a, 27-dihydroxycholesterol (7a27-OHC)
7a, 24(s)-dihydroxycholesterol (7a24S-OHC)
7b, 25-dihydroxycholesterol (7b25-OHC)
7b, 27-dihydroxycholesterol (7b27-OHC)
25-hydroxycholesterol (25-OHC)
27-hydroxycholesterol (27-OHC)
24(S)-hydroxycholesterol (24S-OHC)
Glutathione reduced (GSH) / Glutathione oxidized (GSSG)
Cysteine (Cys) / Cystine (CySS)
Homocysteine (Homocys) / Homocystine (HomocySS)
