ZOETE Lab
Our focus
The laboratory specializes in the development of computer-aided algorithms, programs and databases for molecular modelling, protein engineering and drug design, with applications in oncology, notably in immunotherapy of cancer. We also provide support molecular modeling support to the Molecular Tumor Board of the Lausanne University Hospital.
Our projects
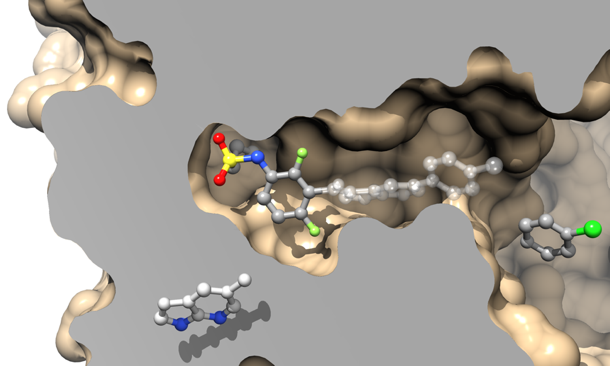
Computer-aided drug design
We are also developing programs for computer-aided drug design. For instance, we are currently designing a novel knowledge-based algorithm to evaluate the relevance of virtual ligand-protein binding modes calculated by docking software, independently of the scoring function of these programs. We are also optimizing existing molecular similarity estimators for use in ligand-based virtual screening. Based on this technology we are designing a novel estimator of affinity levels of small drug-like molecules for a large number of different protein targets. These approaches are used in several drug design projects, in collaboration with academic groups or industrial partners.
Computer-aided protein engineering
The laboratory is developing structure-based computer-aided approaches for protein engineering, notably to optimize the recognition between two macromolecules of interest. In collaboration with groups from the Department of oncology UNIL CHUV, we are also applying these approaches to the design of new immunotherapies against cancer, by engineering protein sequences that could lead to a better recognition of tumor cells by T-cells, and to a stronger T-cell activity.
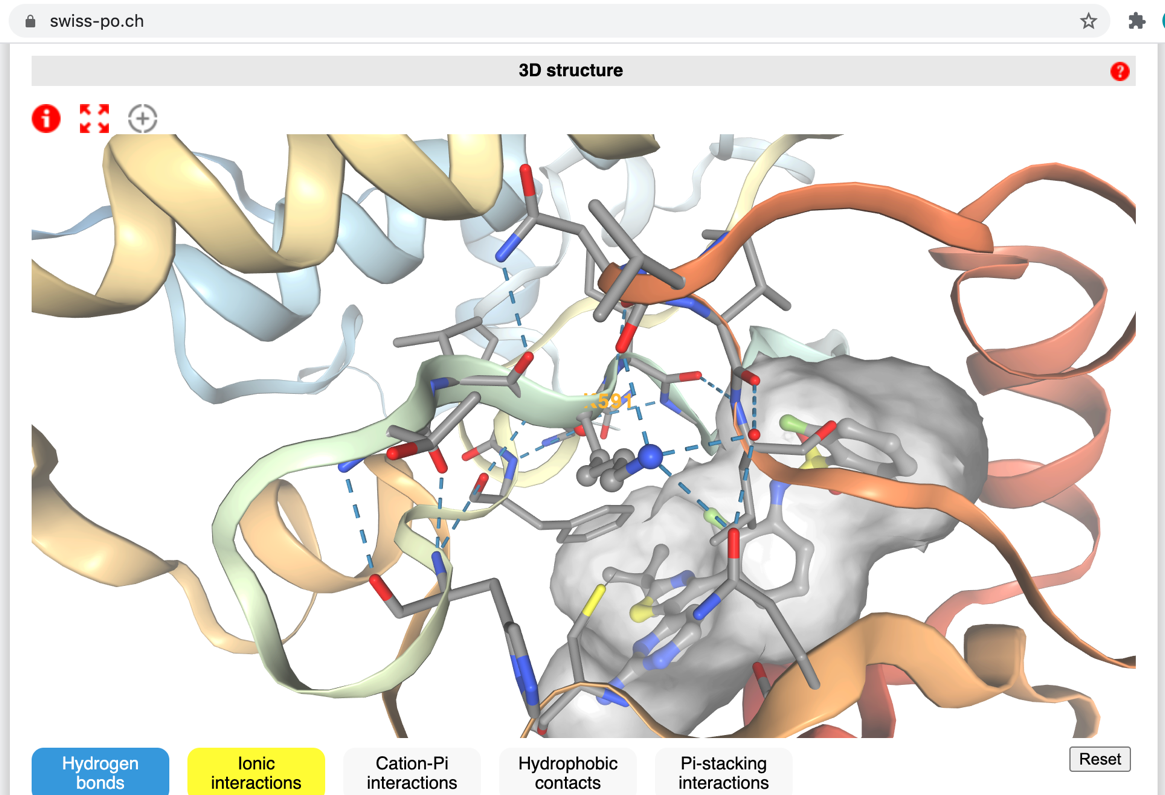
Rapid estimation of mutation impact on protein structure and activity
We are developing several approaches and tools to help predicting the impact of mutations on protein structure and activity.
The objective of these new approaches is, among others, to contribute facilitating the choice of the most appropriate cancer treatment as a function of the particular genetic alterations found in the cancer cells of the patient, or to select patients who could be enrolled in particular clinical trials. In addition to these theoretical developments, we are also designing web tools, such as Swiss-PO.ch, which could be used in Molecular Tumor Boards to simplify and accelerate the prediction of the possible effects of newly encountered mutations. This includes the development of a rapid machine-learning based estimator of the potential impact of a given mutation based not only on the nature of the mutation, the sequence conservation across species of this particular residue, but also on 3D structural information, when available.
These approaches are used on a weekly basis in the context of the Molecular Tumor Board of the Réseau Romand d’Oncologie, and in projects with pharmaceutical companies.
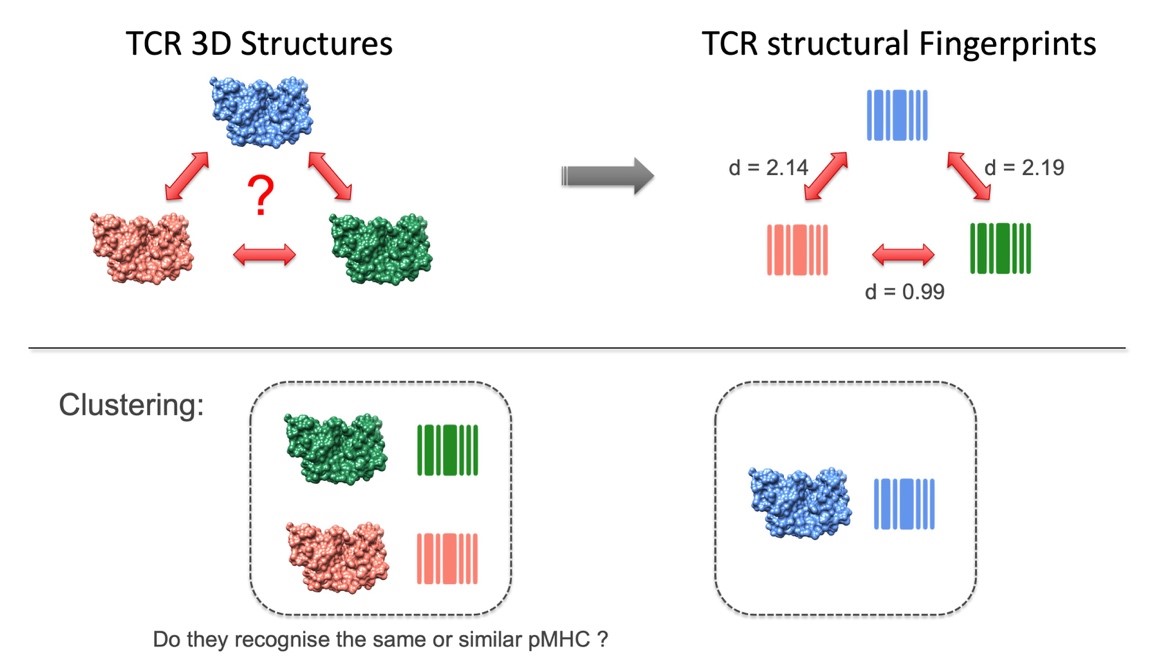
Analysis and prediction of T-cell receptor repertoires
Our group is developing several approaches for the clustering and analysis of TCR repertoires found in patients. These approaches make use of the physico-chemical properties and solvent accessibility of key TCR residues. Another, more novel, class of approaches is being developed which uses structural fingerprinting of TCR surfaces to calculate distances or similarities between TCRs and ultimately to cluster them. Both the physics-based and the fingerprint-based distances are developed so that the similarity principle often used in drug design, and which states that similar small molecules are likely to share similar bioactivities, also applies to protein-protein association and notably to TCR-pMHC binding. These new and rapid similarity estimators could also be used by machine-learning approaches to predict TCR specificities in the future, with possible clinical applications.
KEY PUBLICATIONS

-
Krebs, F; Zoete, V*; Trottet, M; Pouchon, T; Bovigny, C; Michielin, O. Swiss-PO: a new tool to analyze the impact of mutations on protein three-dimensional structures for precision oncology. Npj Precision Oncology. 2021, 5(1):19.
-
Krebs, FA; Gérard, C; Wicky, A; Aedo-Lopez, V; Missiaglia, E; Bisig, B;Trimech, M; Michielin, O; Homicsko, K; Zoete, V. Trametinib Induces the Stabilization of a Dual GNAQ p.Gly48Leu- and FGFR4 p.Cys172Gly-Mutated Uveal Melanoma. The Role of Molecular Modelling in Personalized Oncology. International Journal Of Molecular Sciences. 2020, 21(21):8021.
-
Röhrig UF, Reynaud A, Majjigapu SR, Vogel P, Pojer F, Zoete V. Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (IDO1). J Med Chem. 2019, 62(19):8784-8795.
Meet all the Zoete Lab Members.
|
Funding |
|
|
Affiliations |
|
|
Links |








