About me
I’m an ecologist by training, an evolutionary biologist by trade rapidly evolving into an Evo-Devo scientist.
When I started doing research, I was fascinated by the interaction between the environment and organisms. To be honest, as many students of the Natural Sciences, I hoped to travel to exotic places, like the famous naturalist explorers of the 19th century, and study the incredible biodiversity of those ecosystems. With that goal in mind, I obtained a fellowship and designed my own PhD project to make it happen! That’s how I found myself studying the amphibian community of Mount Kilimanjaro in Tanzania.
After chasing frogs up and down the roof of Africa, I joined Wolfgang Wüster’s lab to investigate the causes and mechanisms of variation in venom composition in the Mohave rattlesnake (Crotalus scutulatus). And that’s how I discovered the wonderful world of animal venoms!
When I’m not doing venomous stuff, I’m either training for endurance triathlon, snow boarding, hiking or scuba diving… anything that gives a good level of adrenaline and stamina :)
About my Research
Venomous animals are found throughout the entire tree of life, where organisms have independently evolved sophisticated apparatuses to produce and deliver potent biochemical weapons. The evolutionary acquisition of venom remodels the predator-prey interaction from a physical to a biochemical battle enabling small animals to defeat larger organisms. Acquiring such weaponry involves evolving a “factory” for venom production including, for instance, exocrine glands, ducts muscular connections and innervations, as well as a delivery system such as fangs, stingers, harpoons, forcipules among others.
Evolution of venom regulation in Neogastropoda
Within the framework of my Marie Curie project EVER, I use cone snails as a model system to investigate how the venom gland evolved the ability to secrete toxins.
Recently, in collaboration with Rob Waterhouse’s group, we showed (Zancolli et al. 2022 PNAS) that various pathways and regulatory proteins have been repeatedly adopted for venom production across animals as distance as spiders and snakes (Fig. 1). However, we also observed several lineage-specific trends, possibly reflecting the different developmental origins of venom glands. So how did venom gland transcriptome evolve?
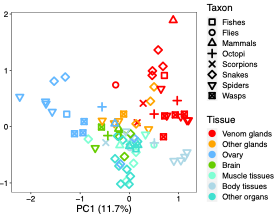
Fig 1. PCA showing venom glands (red) of different lineages
clustering together. From Zancolli et al. 2022.
Fact is – in most lineages, we don’t know which structure is homologous to the venom gland. One exception though are cone snails, a group of voracious predatory marine snails. Cone snails are the only neogastropods with a sophisticated apparatus to secrete and deliver venoms so potent to kill a fish in a matter of seconds (check out this video). The other clades possess a variety of esophageal glands (Fig. 2) homologous to the venom gland. In this project, I’m comparing gene expression patterns of these glands and other body tissues to understand whether novel genes or the differential orchestration of pre-existing genes contributed to the evolution of the venom system.
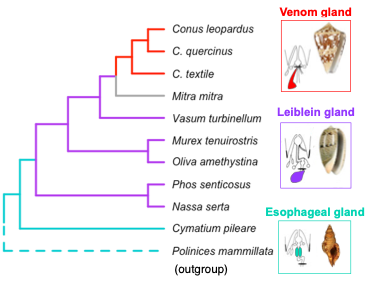
Fig 2. Neogastropoda species investigated and their esophageal glands.
Preliminary results show that many genes selectively expressed in the venom glands are found exclusively in the cone snail clade; additionally, many venom-gland specific orthogroups are larger in venomous species suggesting that novel genes as well as duplication and neofunctionalization of pre-existing genes contributed to the evolution of the specialization in venom production.
This project wouldn’t be possible without the collaboration with Maria Vittoria Modica, Nicolas Puillandre, and the MNHN New Caledonia expedition.
Spatial transcriptomics of cone snail venom system
In this project, funded by my Marie Curie fellowship and a Unil ProFemmes grant, I collaborate with Aida Verdes to study gene expression patterns across the venom gland of the Mediterranean cone snail, Lautoconus ventricosus using a novel and powerful spatial technology.
Cone snails not only have evolved a sophisticated apparatus to secrete venom, but they also acquired the ability to deploy different sets of toxins to defend themselves or for predation (Fig. 3). These toxins are differentially expressed and synthesized along the venom duct, with defense-evoked venom produced in the proximal region (P) whereas predation-evoked toxins are synthetized in the distal region (D). Structural differences are also observed along the venom duct. Here, we use the 10X Genomics Visium slide to investigate the structural and molecular basis underlying functional specialization of venom and its secretory tissue.
How can we fit a 10 cm long duct onto a 6x6mm Visium capture area? Thanks to the skills of our collaborator Manuel Tenorio, we were finally able to place the venom gland as a Swiss roll, check it out in Fig. 3!
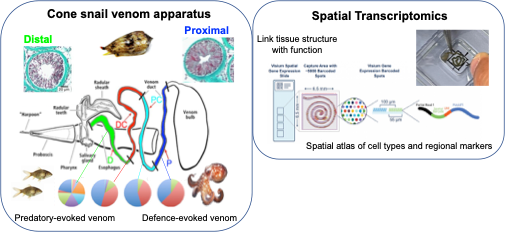
Fig 3. The venom apparatus of cones snails is composed by a long, convoluted duct, a darting harpoon, and a muscular bulb. We are using spatial transcriptomics to investigate the functional and structural specialization along the venom duct and bulb. On the left: from Dutertre et al. 2014 and Hu et al. 2012.
|
Pioneering a new field of research: venom evo-devo In this exciting SNSF-funded project, I collaborate with Yehu Moran and Alistair McGregor to investigate, for the first time, the mechanisms underlying the morphogenesis of the venom apparatus, using the common house spider, Parasteatoda tepidariorum, as a model system. By combining high-definition morphological reconstructions (X-ray synchrotron), spatiotemporal analysis of gene expression dynamics (10X Visium and single-cell RNA-Seq), and genomic manipulations (RNAi) we aim to reconstruct in 3D the development of the venom apparatus and to reveal the key players involved in the emergence and differentiation of a complex, centralized venom system. In doing so, we will establish a new area of venomics: venom evo-devo. |
 |
|
Have Uloboridae lost their venom, for real? In this spontaneous, out of interest, project, I work with Tim Lüddecke and Peter Michalik to verify whether Uloborus plumipes effectively lacks a venom system (the only documentation is a paper from 1931 with a hand drawing). These funny spiders have evolved a peculiar strategy to immobilize and kill their prey – they extensively wrap the prey with silk and cover it with regurgitated digestive enzymes (check out this video here). Whether the prey is killed by suffocation or potential toxins in the digestive fluids or silk is unknown and that is what my Master student Xiaojing Peng is trying to figure out |
 |
Other science related activities
I’m on the Management Committee of the European Venom Network COST Action CA19144 (EUVEN) as country representative for Switzerland and co-leader of working group 4 “Web Resources”. Our goal is to boost communication between research groups and other collectives, provide standards in venom research, and nurture a new generation of venom researchers. Check out our website here.

Giulia Zancolli, PhD
Google Scholar: https://scholar.google.ch/citations?hl=en&user=BXdENsYAAAAJ
OrcID: https://orcid.org/0000-0003-3060-2507
Education
04.2010 – 12.2013: PhD in Integrative Biology
Supervisory committee: Ingolf Steffan-Dewenter, Dieter Mahsberg and Mark-Oliver Rödel.
University of Würzburg, Germany
09.2007 – 09.2009: MSc in Conservation and Animal Biodiversity
University of Turin, Italy
09.2003 – 03.2007: BSc in Natural Sciences
University of Turin, Italy
Employment history
01.07.2019 – present: Marie-Curie Fellow
DEE, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland
Project: “EVER – Evolution of Venom Regulation”. Marc Robinson-Rechavi’s group.
01.12.2018 – 31.12.2019: Bioinformatician and Data Analyst
SIB, Swiss Institute of Bioinformatics, Lausanne, Switzerland
Bioinformatics Core Facility (BCF), Mauro De Lorenzi’s group
01.04 – 31.10.2018: Journal Development Specialist
Frontiers, Lausanne, Switzerland
01.06.2014 – 30.06.2016: Research Officer
Molecular Ecology and Fisheries Genetics Laboratories, Bangor University, UK
Project: “Going with the flow? The genetic basis for snake venom evolution”. PI: Wolfgang Wüster
01.01 – 31.05.2014: Postdoctoral Research Fellow
Department of Animal Ecology and Tropical Biology, University of Würzburg, Germany
01.04.2010 – 31.12.2013 PhD Candidate
Department of Animal Ecology and Tropical Biology, University of Würzburg, Germany
Project: “Amphibian diversity along the slope of Mount Kilimanjaro: from species to genes”.
Approved research projects as PI or co-PI
01.05.2022 – 30.04.2026: SNSF COST Project Grant IZCOZ0_205460
Project: “Pioneering a new field of research: Venom evo-devo”. Role: PI, CHF 278,232
01.01.2022 – 31.12.2023: ProFemmes Grant
EDI Commission, Faculty of biology and Medicine, University of Lausanne
Project: “Regulatory evolution of animal venom system”. Role: PI, CHF 100,000
Prizes, awards, fellowships
01 – 30.12.2021: E-COST EUVEN Virtual Mobility Grant
“Survey on web resources in venom research”, € 1,500
01.06.2019 – Present: H2020 Marie Skłodowska-Curie Individual Fellowship
“EVER – Evolution of Venom Regulation”. € 286,724.16
10.04 – 16.05.2017: European Molecular Biology Organization (EMBO) Short-Term Fellowship
“Mapping an arsenal: molecular cytogenetics of venom”. € 2,636.36
07.2016: Young Investigators Travel Award
Society for Molecular Biology and Evolution. $ 2,000
04.2016: Santander Early Career Researcher Scholarship
Bangor University. £ 3,000
01.01 – 31.05.2014: Career Development Fellowship
Graduate School of Life Sciences, University of Würzburg. € 14,000
08.2013: German Academic Exchange Service (DAAD) Travel Grant “PROMOS”
Graduate Schools, University of Würzburg. € 1,000
06.2012: Universitätsbund Würzburg Travel Grant
University of Würzburg. € 2,000
04.2010 – 12.2013: German Research Foundation (DFG) Excellence Initiative Fellowship
PhD Scholarship, Graduate School of Life Sciences, University of Würzburg. € 116,800
Institutional responsibilities
31.01.2022: Master thesis evaluation panel
University of Lausanne, Lausanne, Switzerland
01.02 – 31.03.2017: Scientific Consultant
Scottish Heritage Foundation
01.09.2014 – 30.06.2016: Molecular Ecology Seminar Organiser
Bangor University, Bangor, UK
Supervision of junior researchers
01.02.2022 - Present: Master thesis supervisor
University of Lausanne, Lausanne, Switzerland
01.10 – 15.12.2020: First-Step project supervisor
University of Lausanne, Lausanne, Switzerland
Teaching activities
01.10.2014 – 30.06.2016: Molecular biology techniques
Bangor University, Bangor, UK
Membership in boards and Reviewing activities
01.10.2020 – Present: Management Committee member of EUVEN COST Action (CA19144)
01.12.2013 – Present: Peer-reviewer
15.05.2017 – Grant External Reviewer for the French National Research Agency

